

“Decoding Cancer: Cancer Genome Atlas Director Dr. Jean Claude Zenklusen in Conversation with Museum CEO John Hollar,” June 21, 2017. Produced by CHM Live at the Computer History Museum.
“Some people say, ‘Oh, I have the Monday morning problems’ or the ‘Sunday afternoon blues’ because you have to go to work, but I never have that. I really go to work fully aware of what we are doing and fully aware of the necessity of what we are doing,” said Dr. Jean Claude Zenklusen, director of The Cancer Genome Atlas (TCGA), during a conversation with CHM Live moderator John Hollar.
TCGA is a collaboration between the National Cancer Institute (NCI) and the National Human Genome Research Institute (NHGRI) and is the largest-scale cancer genomics project to date. Its researchers have mapped key genomic changes in 33 different types of cancer, including 10 rare forms of the disease. The organization has also collected 2.5 petabytes of data describing tumor and normal tissues from more than 11,000 patients. This information is publicly available and has been used by thousands of researchers.
On June 21, 2017, TCGA Director Dr. Jean Cluade Zenklusen joined CHM’s John Hollar for a compelling conversation about the intersection of genetics and technology in studying, treating, and preventing cancer.
Understanding a person’s genetics has become crucial to better understanding cancer and, by extension, cancer treatment. On stage, Dr. Zenklusen explained the sequence of events that needs to occur to turn a normal cell into a cancer cell. But first, he began with an explanation about cancer and its link to age.
For the purposes of this conversation, cancer is a genetic disease that often occurs after people reach a certain age. (Dr. Zenklusen explained that not all cancers are genetically based. A majority of childhood cancers, such as leukemia, are caused by hematological malignancies, abnormalities found in the blood. These are rare forms of cancer.) In order for a normal cell to become cancerous, one generally needs to be exposed to cancer-causing agents, such as sunlight, cigarette smoke, or carcinogens at the workplace, over an extended period, or throughout one’s lifetime. And, while exposure to such agents is a common cause, no one is unfortunately excluded from mutations occurring randomly. Repeated exposure to agents can cause regular genes to mutate in the same cell; it takes time for this kind of mutation to occur. Repeated genetic mutations of this kind (that is, mutations in the same cell) can cause a regular cell to transform into a cancer cell.
Dr. Jean Claude Zenklusen explains the cycle that has to occur in order to turn a normal cell into a cancer cell...
A cancer cell, unsurprisingly, is specific to the person who carries it because of the series of events that causes the transformation. Dr. Zenklusen cites breast cancer, which is actually five different diseases, each with their own biological makeup, unique to the person. Extensive data generated by TCGA provides researchers with a better understanding of the genetic alterations that have taken place in a cell. By analyzing this data, researchers now have a better idea of the biological characteristics that define certain cancer types, giving them a better insight into treatment.
TCGA may be the largest-scale genomics project to date, but Dr. Zenklusen says that the human imagination is just as important as automation. According to Dr. Zenklusen, TCGA would not be able to produce such high quantities of data if the process was a manual one. Automated technology has freed up teams of medical professionals to do the more important, creative work of analyzing and interpreting the data—“at least for the foreseeable future,” Dr. Zenklusen quips.
Dr. Jean Claude Zenklusen talks automation versus human imagination in the largest-scale genomics project to date, The Cancer Genome Atlas (TCGA)...
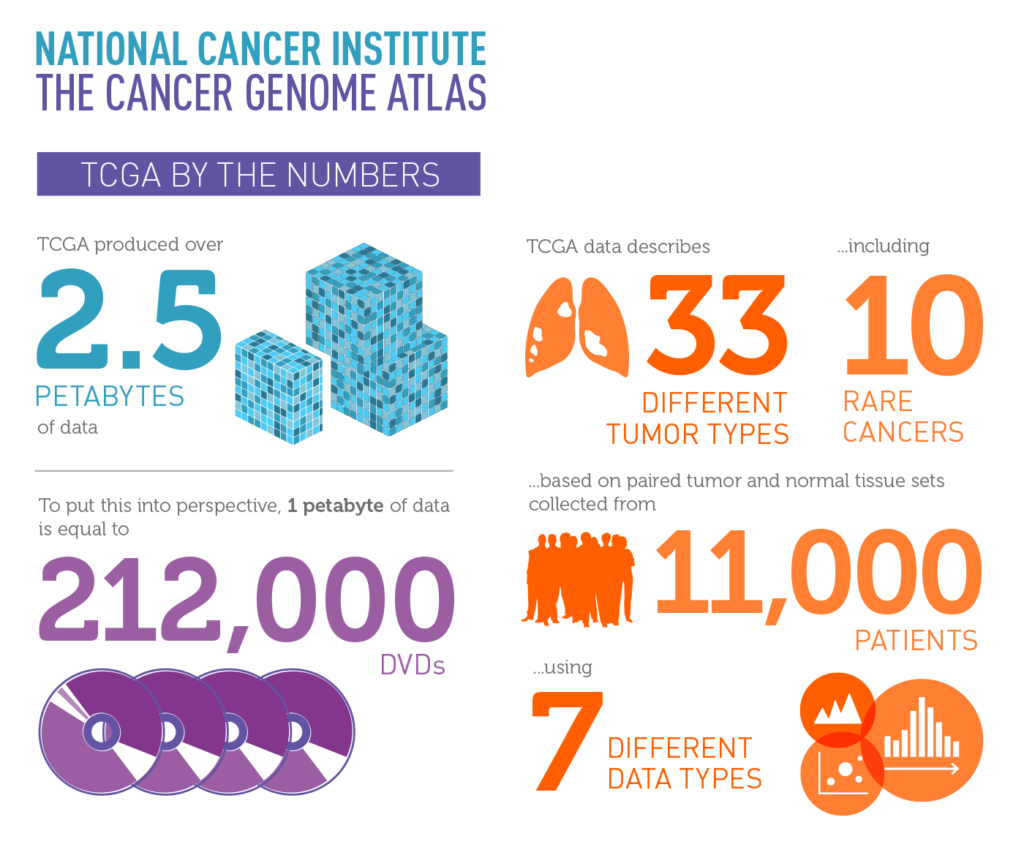
Various statistics about the National Cancer Institute's Cancer Genome Atlas.
An ongoing challenge for TCGA has been accessibility. In June 2016, TCGA launched its first iteration of the Genomic Data Commons, a comprehensive database and repository housing all 2.5 petabytes of its findings. The drawback: in order to view this information, files had to be downloaded, a task that Dr. Zenklusen candidly admitted was simply not feasible. A year later, in June 2017—shortly after this talk—a second iteration of the Genomic Data Commons was launched. This latest version is equipped with visualization tools and advanced search and comparison functions. Now, everyone, including the general public, scientific researchers, and doctors around the world, can access and view information directly on the site, for further analysis.
Dr. Jean Claude Zenklusen explains the scope of The Cancer Genome Atlas (TCGA) and the challenge of making TCGA’s data accessible...
In response to an audience question about where someone with an interest in computer science can go to learn more information about cancer research, Dr. Zenklusen championed the National Cancer Institute’s website and, then, with a not-so-subtle raise of his hand, added, “Find someone who is willing to talk your ear off.” He goes on to share his introduction to computer science nearly 10 years ago during a project called REMBRANDT—a data portal for the Glioma Molecular Diagnostics Initiative he was directing at the time—that transformed how he thought about medical analysis and collaboration.
Why Dr. Jean-Claude Zenklusen thinks collaboration across all scientific fields is the key to learning..
Dr. Zenklusen is 50. He says he’ll retire in about 20 years, and as he looks toward his future and the future of cancer and genomics research, he believes that, while there will never be a single cure for cancer, there will be many tumor types that are no longer life-threatening.
Why Dr. Jean-Claude Zenklusen says there will never be one cure for cancer...
“Decoding Cancer: Cancer Genome Atlas Director Dr. Jean Claude Zenklusen in Conversation with Museum CEO John Hollar,” June 21, 2017. Produced by CHM Live at the Computer History Museum...
The CHM Live team caught up with Dr. Zenklusen to ask him some rapid-fire questions about himself. Here’s what he had to say.
CHM Live: Who are you following on social media?
Zenklusen: I am too old to do social media. Younger generations do live by it, but I actually have a real life to attend to outside the media sphere.
CHM Live: What is one piece of technology you can’t live without? Zenklusen: My iPod (or really my music library). [I] Have about 27 days worth of music (mostly classical, opera, and jazz). I can only concentrate at work if I am plugged unto some kind of music.
CHM Live: What is the first thing you do in the morning?
Zenklusen: Get coffee and check the news on TV. I know, old-fashioned.
CHM Live: What’s the best piece of news you’ve ever received?
Zenklusen: Both pregnancies of my two boys.
CHM Live: What’s your favorite book?
Zenklusen: I have many. The Tristia by Ovid is one. Poignant description of exile and longing. Resonates with me because I have been a perennial immigrant all my life. Love Molière, find it very entertaining. Tolkien and the Dune series [by Frank Herbert] as well. Latest love is the Song of Ice and Fire series (a.k.a., Game of Thrones); I hope R. R. Martin will actually write the last books!
CHM Live: What’s on your playlist?
Zenklusen: Mostly opera, classical, choral, and jazz. Some pop and rock as well (my guilty pleasure is ABBA).
CHM Live: What’s the best part of your job?
Zenklusen: Meeting new people, connecting people from different fields so they can create bigger stuff than they can do alone.
CHM Live: What’s a project or activity outside of work that you’re currently passionate about?
Zenklusen: Gardening. A life-long passion that makes my troubles melt away.
CHM Live: What did you want to be when you grew up?
Zenklusen: Originally an architect. Then I had my first encounter with chemistry and I was hooked. Still love good design, and I appreciate it when I travel to new places.
CHM Live: What’s your favorite piece advice?
Zenklusen: Given by my PhD advisor (Claudio Conti): “Take care to cultivate your family relations; diplomas, awards, and accolades are poor companions in your old age.”
CHM Live: What was your first computer?
Zenklusen: Commodore 64. (See it in the Museum’s permanent exhibition, Revolution: The First 2000 Years of Computing.)
CHM Live: Are you a MAC or PC user?
Zenklusen: MAC, for 25 years now.
CHM Live: Describe a moment when you met someone famous and felt “starstruck.”
Zenklusen: Met Gertrude Elion (Nobel Prize in Medicine for development of a host of drugs) at the National Science Student Symposium in Galveston Texas. She was judging posters and presentations, I was presenting as a graduate student. Amazing woman, great scientist and yet it felt as I was talking to my grandmother if she had understood science. I still get a warm feeling every time I think about it.
On display during this conversation was CHM’s Paracel GeneMatcher. The GeneMatcher began as a top secret government project at TRW called the Fast Data Finder (FDF). It was introduced in 1997 and was based on custom application-specific integrated circuit (ASIC) technology to specifically analyze genetic information, including differences of DNA or protein sequences in relation to other genetic sequences. It contained about 27,000 programmable processors and was originally used by the University of Washington’s Institute of Systems Biology for the Human Genome Project. (Contributed by Dag Spicer, Senior Curator.)
Gift of Michael M. Hansen, CHM# 102751910